Unveiling clonotype diversity and repertoire overlap in diseased PBMCs through OmnibusX analysis
Introduction
Understanding the dynamics of T-cell receptor (TCR) and B-cell receptor (BCR) clonotypes is essential for advancing our knowledge of immune system functionality and its role in health and disease. Single-cell RNA sequencing (scRNAseq) combined with TCR/BCR sequencing provides a robust framework for analyzing clonotype diversity and repertoire structure at a granular level.
Clonotype diversity, repertoire overlap, and spectratype analyses are powerful tools for investigating immune responses. These methods shed light on the clonality of T cells, immune memory, and antigen-specific responses, particularly in disease contexts. They also enable the identification of dominant clonotypes that may drive immune responses or contribute to pathogenesis.
In this analysis, we employed the 10k Human Diseased PBMCs (ALL) Freshly Processed dataset, publicly available from 10X Genomics. The dataset was processed using OmnibusX to integrate scRNAseq and TCR/BCR data seamlessly. With a focus on the T cell population, we used OmnibusX’s analytical tools to explore clonotype diversity, repertoire overlap, and clonotype distribution, illustrating how these insights can be achieved with precision and efficiency.
Dataset processing with OmnibusX
The obtained dataset initially contained 13,853 cells. After applying standard quality control (QC) metrics in OmnibusX, 13,594 high-quality cells were retained for analysis.
Preprocessing and integration
OmnibusX streamlined the submission and preprocessing of the dataset using its scRNAseq pipeline. This included quality control, normalization, dimensionality reduction, and clustering to identify distinct cellular populations. The platform also integrated TCR/BCR data with the scRNAseq analysis, linking clonotype information to individual cells, enabling a detailed exploration of immune repertoire characteristics.
Figure 1: Integration of TCR/BCR sequences with scRNAseq data, assigning clonotype information to individual cells.
Automated cell type annotation
Cell types and subtypes were annotated using the OmnibusX Cell Type Prediction tool, which is built on a carefully curated marker gene database. This step ensured accurate and biologically meaningful cell labeling, providing a robust foundation for further analysis of the T cell population.
Figure 2: Automated cell type annotation using the OmnibusX Cell Type Prediction tool. The annotated T cells and B cells accurately align with the detected TCR/BCR sequences.
Focusing on the T cell population
T cells were selected due to their central role in adaptive immunity and their clonotype diversity, which reflects immune system dynamics in health and disease. Investigating their TCR repertoire provides critical insights into clonotype expansion, immune memory, and potential disease-related changes.
For this analysis, we focused on the T cell population containing one or two pairs of TRA and TRB sequences exclusively. Using the sub-clustering functionality in OmnibusX, we isolated this population to ensure a high-confidence dataset for detailed exploration.
Figure 3: Selection of cells containing one or two pairs of TRA + TRB sequences. Cells with multiple TCR chains were excluded, as these may indicate doublets, immature T cells, or assembly errors.
Exploring TCR data with OmnibusX
Clonal expansion analysis
OmnibusX enables seamless clonal expansion analysis by assigning a clonal size (number of cells) to each unique TCR clone from the input data. Clones with a high number of cells are indicative of clonal expansion, reflecting the immune system's response to specific antigens.
Figure 4: Distribution of cells across unique clones, highlighting clonal size.
To statistically assess clonal expansion, OmnibusX performs a binomial test for each clone, providing p-values to evaluate the significance of the expansion. The results are visualized, allowing users to identify and select cells associated with significantly expanded clones. For this analysis, we selected cells with p-values < 0.05 and annotated them as part of clonal expansion for subsequent exploration.
Figure 5: Binomial test results showing p-values for assessing clonal expansion significance.
Figure 6: Annotation of cells with p-values < 0.05 as clonal expansions, enabling targeted downstream analysis.
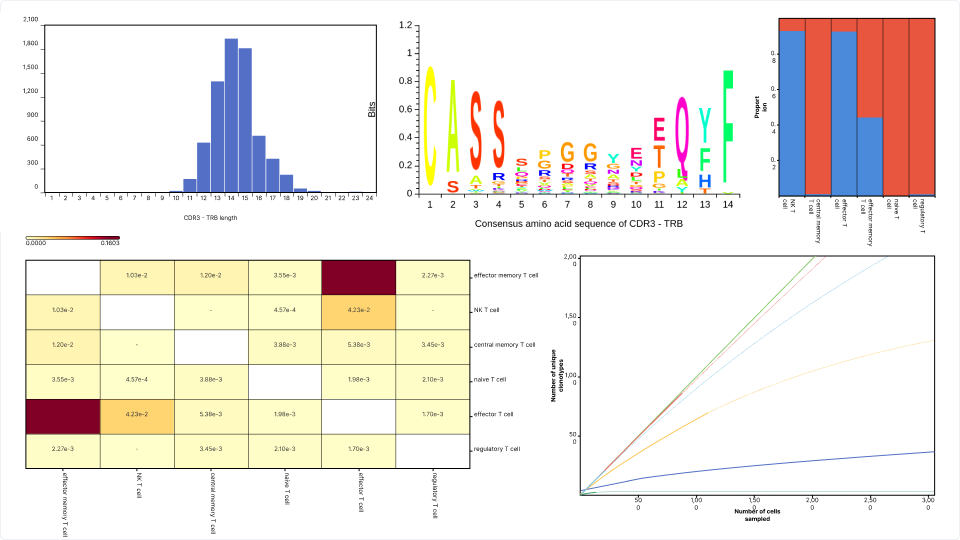
Next, we conducted a composition analysis to explore the relationship between clonal expansion and T cell subtypes, as determined by the OmnibusX Cell Type Prediction tool. The analysis revealed that clonal expansion predominantly occurs in effector T cells and NKT cells. Additionally, approximately half of the effector memory T cell population showed evidence of significant clonal expansion. In contrast, naive T cells, central memory T cells, and regulatory T cells exhibited little to no significant expansion. This indicates the activation of T cells transitioning from a naive to an effector state and the triggering of memory T cells for a faster immune response.
Figure 7: Composition plot showing clonal expansion predominantly in effector T cells and NKT cell populations, along with a subset of effector memory T cells.
By linking clonal expansion with specific T cell subtypes, we identified key populations actively participating in the immune response, such as effector and NKT cells.
Clonotype diversity analysis: Rarefaction curve
Rarefaction curves are essential tools for visualizing and comparing clonotype diversity within cell populations. By plotting the number of unique clonotypes as a function of the number of cells sampled, these curves provide insights into the richness and evenness of clonotypes across subpopulations.
In this analysis, the diversity was quantified using the Hill number with q=1. The Hill number is a framework for measuring diversity, where q determines the sensitivity to species frequencies:
- q=0: Represents species richness, weighting all clonotypes equally, regardless of their abundance.
- q=1: Corresponds to the exponential of Shannon entropy, balancing richness and evenness while avoiding overemphasis on rare or dominant clonotypes.
- q=2: Focuses on dominant clonotypes, giving greater weight to more abundant ones.
We chose q=1 to provide a balanced measure of clonotype diversity, capturing both richness and evenness across the T cell subtypes.
Using OmnibusX, rarefaction curves were generated for five T cell subtypes identified through the OmnibusX Cell Type Prediction tool:
- NK T cells
- Central memory T cells
- Effector T cells
- Effector memory T cells
- Naive T cells
- Regulatory T cells
The rarefaction curves reveal significant differences in clonotype diversity among these subtypes. The diversity decreases in the following order:
- Naive T cells
- Central memory T cells
- Regulatory T cells
- Effector memory T cells
- Effector T cells
- NK T cells
Figure 8: The rarefaction curves with q=1 reveal significant differences in clonotype diversity among T cell subtypes.
The curves provide valuable insights into the biological roles and clonotype dynamics of each subtype:
- Naive T cell: The rarefaction curve for naive T cells rises sharply and continues to grow, indicating a broad repertoire of unique clonotypes. This diversity aligns with their role as precursors for adaptive immunity, ensuring readiness to respond to diverse antigens.
- Central memory T cell: Central memory T cells display moderate diversity, with a plateau indicating a more focused repertoire optimized for responding to previously encountered antigens.
- Regulatory T cell: Regulatory T cells exhibit balanced diversity, reflecting their specialized role in maintaining immune regulation and tolerance.
- Effector memory T cell: The diversity of effector memory T cells is lower, with the curve plateauing earlier, signifying a repertoire shaped by clonal expansion in response to specific antigens.
- Effector T cell and NK T cell: Effector T cell and NK T cell demonstrate the least diversity, as their repertoire is dominated by a small number of clonotypes that expand during immune activation.
These results highlight the spectrum of clonotype diversity, ranging from the extensive, uncommitted repertoire of naive T cells to the highly specialized, antigen-driven clonotypes of effector and effector memory T cells. Understanding these differences is crucial for unraveling the complexity of immune responses. To further investigate the relationship between these subtypes, we next explore repertoire overlap analysis, shedding light on how clonotypes are shared or distinct across T cell populations.
Repertoire overlap analysis
Repertoire overlap refers to the degree of shared clonotypes between two cell populations. It provides valuable insights into how immune repertoires are distributed across different cell types, reflecting their functional relationships and potential lineage connections. Understanding repertoire overlap is particularly important in studying immune responses, as it reveals how T cell subtypes contribute to immunity and antigen specificity.
Calculation of repertoire overlap in OmnibusX
In OmnibusX, repertoire overlap is quantified using Jaccard similarity scores, a metric that measures the fraction of shared clonotypes between two populations relative to their combined repertoire sizes. The score ranges from 0 to 1, with higher values indicating greater overlap. For this analysis, the overlap was calculated separately for CDR3-TRA and CDR3-TRB sequences, capturing the shared repertoire at both TCR alpha and beta chains.
Figure 9: Repertoire overlap of CDR3-TRA sequences across T cell subtypes, quantified using Jaccard similarity scores. Higher scores indicate greater sharing of clonotypes between subtypes.
Figure 10: Repertoire overlap of CDR3-TRB sequences across T cell subtypes, highlighting the extent of shared clonotypes in the TCR beta chain repertoire.
The analysis revealed the following patterns of repertoire overlap among the six T cell subtypes:
- The highest overlap was observed between effector T cells and effector memory T cells, with a Jaccard score of 0.16 for CDR3-TRA and 0.14 for CDR3-TRB. This substantial overlap reflects their close functional and developmental relationship, as effector memory T cells are often derived from effector T cells during immune responses.
- All other pairwise comparisons showed very low Jaccard scores, with most values falling below 0.01. This indicates that the clonotypes in these subtypes are largely distinct, consistent with their specialized roles in the immune system.
These findings emphasize the close relationship between effector and effector memory T cells while showcasing the unique repertoire profiles of other subtypes. To gain a deeper understanding of the T cell population's clonotype distribution and structural diversity, we next turn to spectratype analysis, which provides a detailed view of the length distribution of CDR3 sequences within each subtype.
Spectratype analysis
Spectratype analysis examines the distribution and structure of T cell receptor (TCR) clonotypes by focusing on the lengths and sequences of the complementarity-determining region 3 (CDR3), the most variable and antigen-specific region of the TCR. This analysis provides critical insights into clonotype diversity, immune repertoire skewing, and responses to antigens.
Visualization and interpretation in OmnibusX
OmnibusX offers a comprehensive set of tools for conducting spectratype analysis. Users can:
- Visualize consensus sequences: OmnibusX generates weblogo plots for CDR3 sequences across all TCR chains present in the dataset (e.g., CDR3-TRA and CDR3-TRB). These plots reveal conserved and variable regions within the sequences, enabling researchers to identify patterns of amino acid usage that may indicate antigen-specific signatures.
- Examine chain length distributions: The application provides detailed histograms showing the distribution of CDR3 lengths and the corresponding number of cells associated with each length. This helps researchers identify shifts in repertoire structure, such as expansions or contractions of specific clonotypes.
- Inspect detailed sequences: For each clonotype, OmnibusX allows users to explore the detailed CDR3 sequences, offering a granular view of the repertoire at the sequence level.
Figure 11: Spectratype analysis of CDR3-TRA sequences, showing the length distribution of CDR3 regions across the T cell repertoire along with the consensus sequence.
Findings in the diseased PBMC dataset
Using these tools, the diseased PBMC dataset reveals a diverse distribution of CDR3 lengths across TCR chains, with clear subtype-specific patterns. For example, effector T cells and effector memory T cells may exhibit more skewed distributions, potentially reflecting clonal expansion in response to antigens. In contrast, naive T cells tend to show broader, more balanced distributions, consistent with their role in maintaining a diverse immune repertoire.
Research approaches enabled by spectratype analysis
Spectratype analysis in OmnibusX provides researchers with a foundation for various investigative approaches, such as:
- Identifying antigen-specific clonotypes: Conserved motifs in weblogo plots could guide searches for clonotypes associated with specific diseases or antigens.
- Tracking clonal expansions: Length distributions and clonotype frequencies can help monitor immune responses in infection, vaccination, or cancer.
- Exploring repertoire skewing: Comparative spectratype analyses across subtypes or conditions can reveal shifts in immune dynamics, such as those induced by disease or therapy.
By presenting detailed and accessible spectratype data, OmnibusX equips scientists with the tools to investigate TCR repertoire characteristics without bias, leaving interpretation and hypothesis generation open to user discretion.
Conclusion
In this analysis, we explored the TCR repertoire of diseased PBMCs using OmnibusX, demonstrating its powerful and intuitive tools for analyzing single-cell RNA sequencing data paired with TCR/BCR information. Key findings included:
- Clonotype diversity: Rarefaction curves highlighted the wide spectrum of clonotype diversity across T cell subtypes, from the extensive repertoire of naive T cells to the specialized, antigen-driven clonotypes of effector T cells.
- Repertoire overlap: Jaccard similarity scores revealed significant overlap between effector and effector memory T cells, reflecting their close functional relationship, while most other subtype pairs exhibited minimal overlap.
- Spectratype analysis: OmnibusX provided comprehensive visualization of CDR3 sequence distributions, consensus motifs, and sequence-level details, offering insights into TCR structure and repertoire skewing.
These analyses illustrate the flexibility and depth of OmnibusX in uncovering meaningful immune repertoire dynamics, all within a streamlined, user-friendly interface. By enabling researchers to focus on their data rather than technical challenges, OmnibusX facilitates impactful discoveries in immunology and related fields, such as infection, cancer, and autoimmune diseases.
We invite you to explore OmnibusX for your own datasets and leverage its robust analytics to unlock new insights into immune system complexity. The tools are available for download with a 2-month free trial at https://omnibusx.com/apps, providing you with an opportunity to experience its capabilities firsthand.
